User Tools
Sidebar
Table of Contents
Scientific applications, grid computing
This project proposal aims at providing efficient workflow management systems to handle and process large amounts of scientific data on large scale distributed infrastructures such as grids. The target applications in the area of life sciences applications have to deal with distributed, heterogeneous, and evolving large scale databases, to represent complex data analysis procedures taking into account the medical or biological context, and to exploit CS tools to design at a low cost scientifically challenging experiments with a real impact for the community. Two applications are directly considered: one addresses in silico drug discovery while the second focuses on computer aided analysis of cardio-vascular images.
Drug discovery
In silico drug discovery is one of the most promising strategies to speed-up the drug development process. Virtual screening is about selecting in silico the best candidate drugs acting on a given target protein. Screening can be done in vitro but it is very expensive as they are now millions of chemicals that can be synthesized. If it could be done in silico in a reliable way, one could reduce the number of molecules requiring in vitro and then in vitro testing from a few millions to a few hundreds. Virtual screening requires a complex analysis with multiple steps such as molecular modelling, docking and molecular dynamics. Recently, deployment projects of in silico docking on grids emerged in the perspective to reduce cost and time. We propose in this project to further develop and deploy a grid-enabled virtual screening workflow.
Documents
Implementation of the workflow
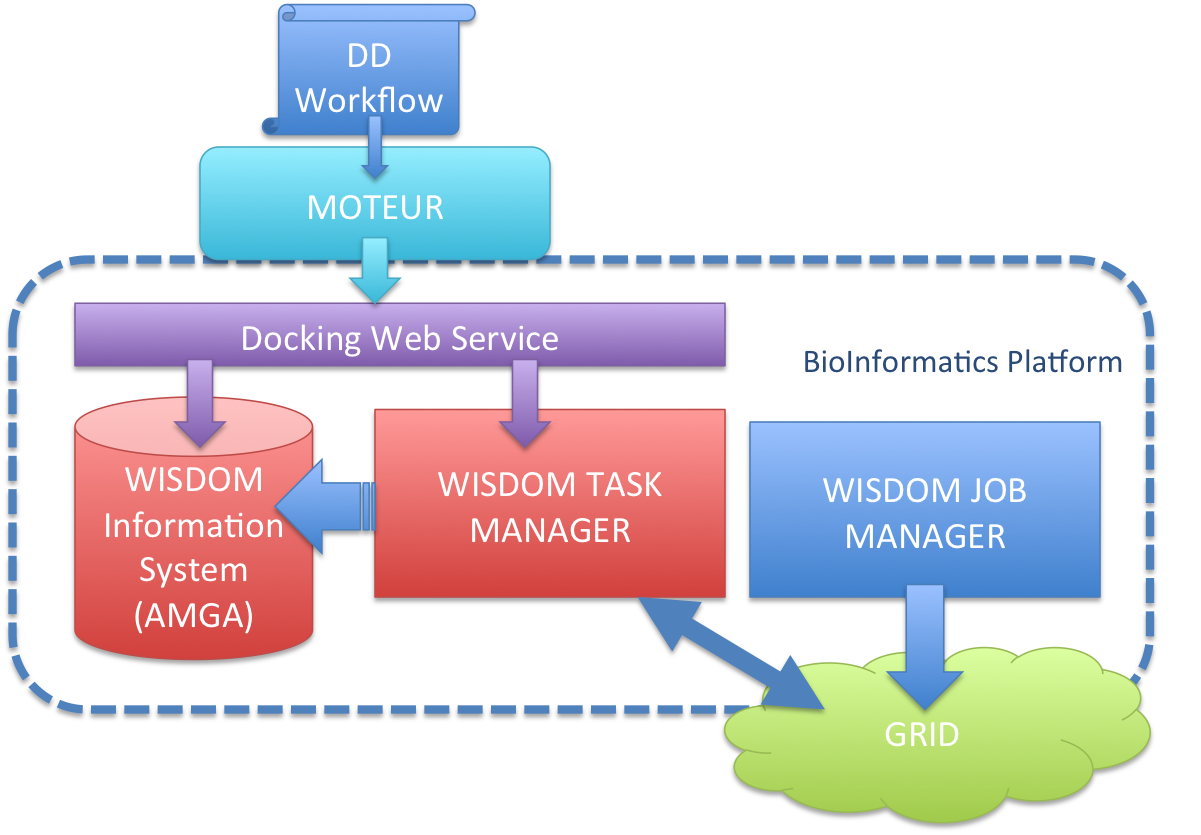
The drug discovery workflow is implemented to be run with the WISDOM environment through the MOTEUR workflow enactor. The following figure shows the overall architecture of the application :
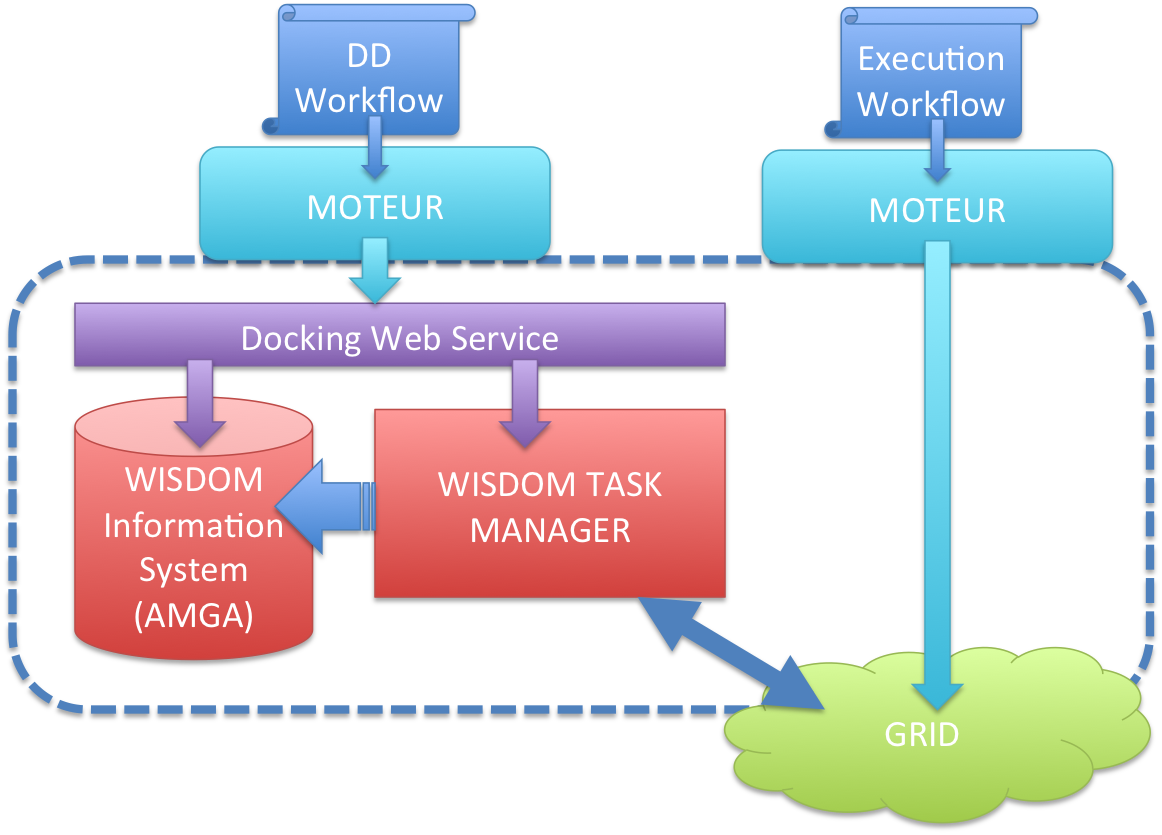
On this figure the grid jobs are managed through the WISDOM JOB MANAGER, but we have another implementation that use a MOTEUR workflow to submit the grid jobs :
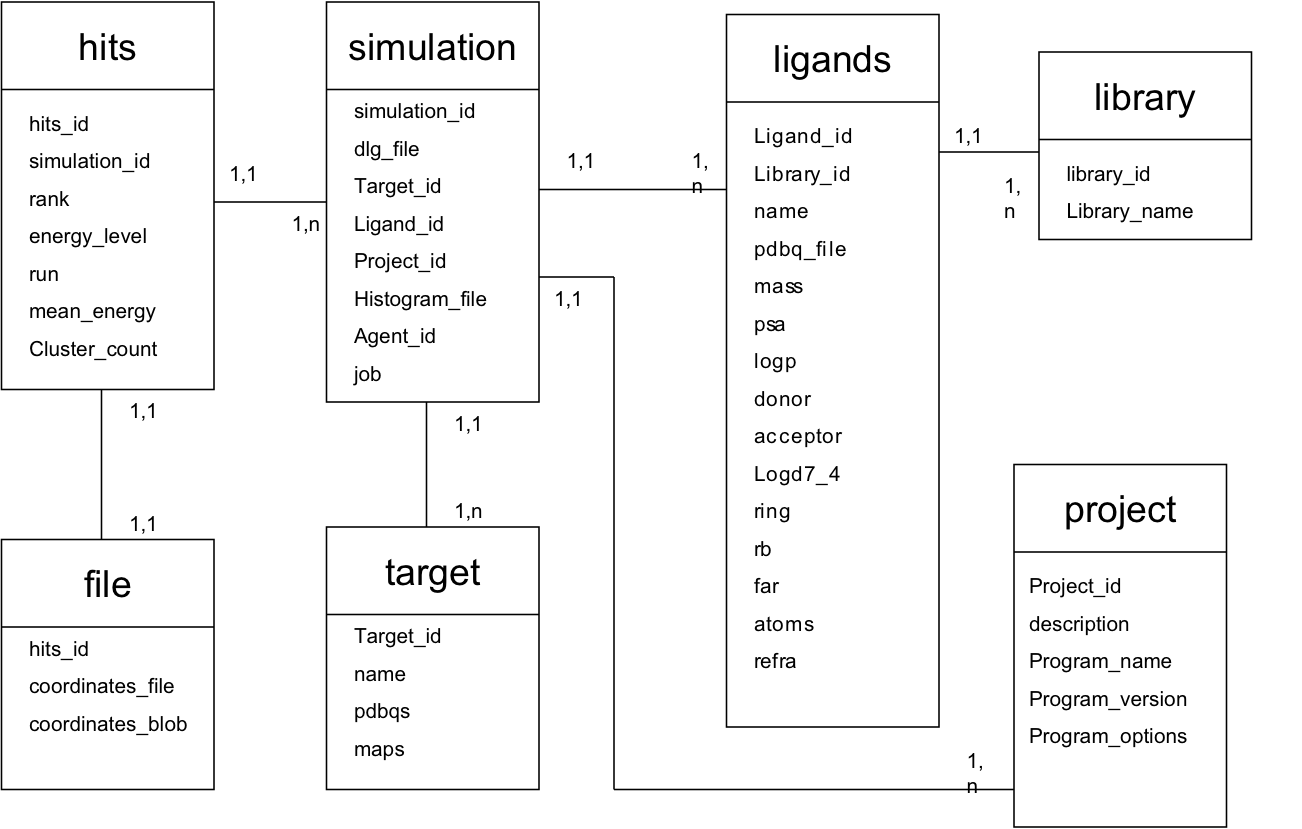
The jobs that are submitted on the Grid will fetch the docking task from the WISDOM TASK MANAGER. The docking web service is managing the creation of tasks and corresponding information in the WISDOM information system using AMGA. This information system contains all the information to run a docking simulation, including, target information, ligand information and docking parameters. The docking database follows the following schema :
The docking parameters are stored in the project table, Under program_options attribute. It is actually a string pointing to the Logical Filename of the parameter file for autodock on the grid.
Future Data Challenge
Last months, WISDOM (Wide In Silico Docking on Malaria) approach has been successfully validated in biochemical laboratories. Using EGEE grid to speed up the calculations, few active compounds against malaria target were highlighted from million-compound databases. This success leads us to expand this approach to other diseases, improving the biological workflow and the grid performances.
Diabetes mellitus is one of the most serious, chronic diseases that is developing along with an increase in both obesity and ageing in the general population. One of the therapeutic approaches for decreasing post-prandial hyperglycemia is to retard absorption of glucose by the inhibition of carbohydrate hydrolyzing enzymes, for example alpha-amylase in the digestive organs. Human Pancreatic Alpha Amylase (HPA) plays a crucial role in the digestion of dietary starch, inhibition of its activity has potential therapeutic value in the treatment of diseases such as diabetes or obesity. Despite the considerable interest in alpha-amylases, there are relatively few other known inhibitors of this group of enzymes.
Proteinaceous inhibitors are generally very potent with typical KIs in the nM range (Feng et al., 1996; Machius et al., 1996; Franco et al., 2000) and synthetic peptides mimicking such inhibitors have also been reported (Sefler et al., 1997). In addition, different carbohydrate inhibitors in the mM (Uchida et al., 1994) and μM (Kim et al., 1999) ranges are known. Considering side effects of carbohydrates inihibitors, we will attempt to discover new non-peptide/carbohydrates towards alpha-amylase in the basis of structure-based virtual screening.
Through the 31 structures are relative to Human Pancreatic alpha amylase available, in the Protein Data Bank (PDB), 3 targets are chosen for virtual screening: 1bsi, 1xcx and 1u2y. PDB contains many anomalies ranging from proteins with small deviations from normal geometry, to structures that fit their submitted experimental data very poorly. For these reasons, within the framework of Embrace european project, a procedure of PDB re-refinement was setted up (JOOSTEN R. and al, Re-refinement of all X-ray structures in the PDB, submitted to Proteins). Re-refined form of 1u2y pdb was consider in our experiment and called 1u2y_ref.
Chembridge drug-like database containing 450,000 compounds is filtered according to key properties of absorption, distribution, metabolism, excretion and toxicity (ADME/tox) that should be considered early in pharmaceutical development. After filtering 300,000 chemicals are still present and used for screening.
The in silico workflow which we employed starts with a total of 1,000,000 dockings (Autodock) to evaluate the binding energy between a target and a ligand, then about ten thousands best compounds resulted from docking will be refined by Molecular Dynamics (Amber 9) to further address the essential parameters like electrostatic solvation which are ignored during the docking process. Both steps are deployed on grid nodes using the Wisdom Production Environment.
Phylogeny Application
The gINFO application represents an automated phylogeny workflow that is run regularly, once per day, to compute phylogenetic trees from virus sequences (see g-info production site). The idea behind it is to use the grid resources to automatically compute and process the new sequences data made available daily as soon as they are published. The gINFO as been implemented in two ways, one hardcoded workflow (which is in production now) that has a predefined script that is used to run the workflow with workflow specific services, and one softcoded workflow, using Moteur2 workflow engine to execute it using non-specific services. Both implementations are fully based on the WISDOM production environment (WPE). The WPE is used to handle the jobs on the grid and maintain constantly some jobs running, while handling all the tasks created through the workflow execution.
- A complete overall documentation can be found here:
http://auvergridpfmanager.univ-bpclermont.fr/ginfo/gINFO_Documentation_Version_finale.pdf
- The code and scripts of the hardcoded (script-based) version can be found here:
http://auvergridpfmanager.univ-bpclermont.fr/ginfo/Workflow_Hard_Scripts/
- The code and scripts of the soft-coded (GWENDIA-enabled) version can be found here:
http://auvergridpfmanager.univ-bpclermont.fr/ginfo/Workflow_Soft_Scripts/
- The WISDOM software packages that are used by the services are available here:
http://auvergridpfmanager.univ-bpclermont.fr/ginfo/wisdom_Services/
These packages are used by the wisdom production environment, and need to be stored at the appropriate location to be executed properly by the WPE. They are useful to understand and see what is happening on each computing node, and how each step of the workflow is executed on the grid. While each service used in the soft-coded implementation can be called independently and generically for any workflow, the hardcoded versions can not be used in a different context since they submit the next step to go in the workflow during their execution, so only the first step has to be submitted and the whole workflow with eventually be enacted whereas “soft” versions need a proper workflow definition and execution by a workflow engine. In our case the workflow is written in gwendia langage and executed by Moteur2.
Cardiac imaging
Cardio-vascular diseases are the leading cause of mortality in our countries. Medical imaging techniques have considerably evolved and now offer a privileged way for studying pathological vessels and hearts (anatomy and functions). However, the multiplicity of examinations results in a huge amount of imaging data which is difficult to fully exploit for the physicians. Therefore, new automatic analysis tools are being developed and results in quantitative parametric representations that ease interpretation and diagnosis. Such tools might be complex combinations of elementary tasks and more sophisticated computer demanding algorithms. In the context of this project, we are interested in using workflows to speed up the design of new cardio-vascular image processing chains and to ease setting up evaluation protocols of such processes on large databases by combining processing tasks. For example, one of the objectives of processing chains will be to automatically extract heart cavity volumes and motion indices directly from 3D dynamic image acquisitions in Magnetic Resonance Imaging.